Optimized Multi-Core Parallel Tracking for Big Data Streaming Applications
Volume 6, Issue 3, Page No 286-295, 2021
Author’s Name: Doaa Ahmed Sayeda), Sherine Rady, Mostafa Aref
View Affiliations
Ain Shams University, Faculty of Computer and Information Sciences, Cairo, 11865, Egypt
a)Author to whom correspondence should be addressed. E-mail: doaa.ahmed74@yahoo.com
Adv. Sci. Technol. Eng. Syst. J. 6(3), 286-295 (2021); ![]() DOI: 10.25046/aj060332
DOI: 10.25046/aj060332
Keywords: Data streams, Spark, Sliding window, K-means++ clustering, Parallel processing, Map-reduce framework
Export Citations
Efficient real-time clustering is a relevant topic in big data streams. Data stream clustering needs necessarily a short time execution frame with bounded memory utilizing a one-scan process. Because of the massive volumes and dynamics of data streams, parallel clustering solutions are urgent. This paper presents a new approach for this trend, with advantages to overcome the main challenges of huge data streams, time, and memory resources. A framework is proposed reliant on a data clustering parallel implementation that divides most recent incoming data streams within a sliding window mechanism to distribute them across a multi-core structure for processing. Every core is responsible for the processing and generation of intermediate micro-clusters for one data partition. The resulting micro clusters are consolidated utilizing the additive property of the micro-cluster data structure to merge those parallel clusters and obtain the final clusters. The proposed approach has been tested on two sorts of datasets: KDD-CUP’99 and KDD-CUP’98. The results show that the proposed optimized parallel window-based clustering approach is efficient for online cluster generation for big data streaming with regard to the performance measures processing time and scheduling delay. The processing time is 1.5 times faster, and the scheduling delay is approximately between 1.3 to 1.7 times less than the sequential implementation. Most important is that the clustering quality is equal to that of the non-parallel implementation.
Received: 12 December 2020, Accepted: 19 March 2021, Published Online: 28 May 2021
1. Introduction
Modern advances in ICT and its utilization in business and life sectors have prompted the quick development of huge volumes of data recognized as big data [1]. The basic attributes of huge data are its dynamics (speed), which indicates that data upon arrival needs prompt processing at varying speediness. While for certain applications, the appearance, and preparation of data can be achieved as offline batch processing, other applications need continuous analysis in real-time for the arriving data [2-4]. Data stream clustering is characterized as the data gathering, considering as often as possible the arrival of new data chunks, while seeking the understanding of examples that may modify after some time [5]. The amount of data coming at high and changeable velocities that assume ordinary clustering algorithms ineffective in terms of meeting real-time requirements, and hence considered incapable of dealing with the requests properly [6].
Data stream clustering includes several difficulties; to meet continuous necessities [7]. The clustering should be executed in a brief period of timeframe with bounded memory using a single-scan process. So, following clusters in the sliding window is possibly an effective way to deal with the restrictions in time and memory [8].
Windowing is one famous handling strategy that is utilized with the data streams. Windowing applied to split data streams into windows, reliant on the time measurement. Exist different sorts of window models for the following changing data streams [7]: 1) Landmark, 2) Damped, and 3) Sliding. In a Landmark window, the window is controlled by a certain time point defined as a landmark and the present. It is utilizing for mining over the whole history of the data streams. Landmark is not fitting for applications where up-to-date data is more significant than obsolete data. In the damping model, weights are determined to data, where higher weights are given to the latest data items and less weight is specified to the outdated data. This implies that the model expects the latest information as more critical than the older data. Sliding window models suppose that the new data is more valuable than outdated data by utilizing a window. The latest occurrences falling inside the window of the data stream are preserved while dispensing with old data. The reason to utilize the sliding window instead of models is regularly basically desired for keeping time and memory resources.
Recently, distributed and parallel algorithms provide solutions for analyzing big data streams in actuality, which is apparent in the more current research works [9]. Parallel solutions and distributed offer various advantages, such as diminishing the time, expanded versatility of arrangements, and suitability for applications of the distributed kind. The currently available multi-core processor commodity computers at afforded costs assist the development of applications in a much easier and reliable way. A computer can contain up to 72 core processors giving high processing power. The inquiry then remains how these available processing onboard resources are utilized for providing optimized solutions for the different real-time in-demand applications.
Most existing parallelism strategies for clustering algorithms cannot give the same clustering quality as the sequential algorithm [1]. The change in clustering quality is impacted influenced by the methods utilized for the data distribution partitioning and results consolidating techniques for the results.
SCluStream [8] is a previously introduced efficient sliding window-based algorithm for clustering data streams considering pre-mentioned data stream difficulties. It works in three phases, wherein the online phase; the most recent data is continuously maintained. The second phase is an expiring phase, where the old data is discarded such that less memory is squandered. The last phase is offline, where the clustering k-means++ algorithm is utilized.
This paper proposes a parallel implementation of the SCluStream algorithm utilizing a multi-core parallel processing framework. The implementation makes the best utilize of the ready cores utilizing a stand-alone platform on a single machine. In the framework, the data set is split into several partitions. Every partition is processed in a separate core, with an equal workload assigned for every partition. Sequential k-means++ is executed next to generate intermediate results (i.e. micro-clusters) for each partition. Eventually, final clusters are consolidated by applying the additive property on micro-clusters, which utilizes the concept of merging the two nearest micro clusters; reliant on the Euclidean distance between micro-clusters.
The organization of this paper is as follows: Section 2 summarizes the related works and existing gaps. Section 3 the spark streaming architecture is explained as the preliminary basis to our big data implementation. Section 4 explains the parallel clustering data streams reliant on the sliding-window and k-means++ parallel algorithm. In section 5 the experimental study and assessments are discussed. In the final section conclusion and future work are clarified.
2. Related work
Clustering huge data volumes require long execution times, especially when talking about dynamic data streams. Many solutions are proposed to beat this issue. Some of them have been done as batch analysis [10-13], while others as streaming analysis [14-19]. Some research enhanced the processing of the algorithm itself by tuning its parameters or modifying the principal framework for the algorithm. Whereas other research opted for the utilization of the parallelism concept, which is often executed utilizing two major strategies. While the first strategy utilizes a network of linked machines, where the clustering algorithm is implemented on a group of computers [10-16]. The second strategy utilizes a single machine with a multi-core processor [11, 18, 19]. In both strategies, the data is distributed among the computers or cores in the first step. Next, comes a consolidation step which should sum up for the correct execution of the clustering algorithm, and which should be roughly as exact as possible when compared to the algorithm executing on a single machine. In the distributed environments, these two aforementioned steps in respective order are commonly identified as the mapping and reducing operations that correspond to the mentioned two steps in respective order. MapReduce [20, 21] is the common framework for effectively writing applications, which process enormous amounts of data in a parallel way.
A new parallel manner for partitioned clustering algorithms reliant on MapReduce is proposed [1]. The target of this optimization is to enhance data distribution on connected nodes and select the best centroids got from every reducer utilizing a GA (genetic algorithm) based results consolidating methodology. In the map phase, the data is distributed by utilizing the maximum distance among the data points of the various partitions. Consolidating the intermediate results acquired from the reducers on a single node utilizing the genetic algorithm to acquire final accurate results.
A common approach for parallel version of DBSCAN is proposed for clustering massive amount of data [11, 13, 18]. A huge dataset is separated into numerous partitions’ dependent on the data dimensions and localized DBSCANs are applied to each partition in parallel through a map phase. The results of each partition are next consolidated in a final reduce phase, which has been performed differently in the three works. A single node tree is at first created for every data point in the dataset by utilizing the disjoint set information structure. Intermediate trees are merged according to the tree-based bottom-up approach after investigating the eps-neighborhood for haphazardly chosen points [11]. A division method named Cost Balanced Partition is utilized to produce partitions with equal capacities and cost-based partitioning (CBP), which determine the partition’s data reliant on the estimated calculation cost [13]. For the merging phase, a graph-based algorithm is used to generate global clusters form local clusters. Complex grid partitioning is used for dividing the data space into several partitions for each dimension to minimize the processing time of DBSCAN in parallel processing [12]. Local DBSCAN is applied on each partition to make local clusters. In the merging phase the overlapping clusters are extracted from spatial clusters in a grid and spatial clusters in grids adjacent to its grid. The taken overlapping spatial clusters are consolidated to create one spatial cluster. This parallel addition meets the prerequisites of scalable execution for managing huge data sets. However, this addition is not appropriate for clustering real-time data. It requires traversing the whole dataset for parallel clustering which infers that its execution time is as yet reliant on the dataset size. Thus, while has a great execution for batch-oriented mode, it isn’t reasonable for high-speed datasets [12, 13].
A presented parallel implementation for clustering high-dimensional data streams in streaming analysis is proposed [14, 15, 17, 22]. pGrid splits the high-dimensional data space into grids that are clustered the grids rather than the raw data [14]. pGrid uses the MapReduce framework. In the mapping phase, the arriving data point is projected onto its matching grid to depend on its dimensions, and thereafter the grid density is varied. In the reducing phase, the grid cells are merged by each dimension and at the end combining overlapping grid cells are combined to generate the global clustering result. PPreDeConStream is proposed with its parallel implementation in that utilized the shared memory model and the online-offline framework [11, 22]. A new data partitioning technique, Fast Clustering (FC) partitioning is applied. The idea of FC is reliant on splitting two 2-dimensional spaces into four sub-cells until a threshold is obtained (i.e. a threshold on the point’s number in a cell) [15]. Then, the cells containing the number of points that are less than this threshold are deleted. The merging phase is based on an overlapping area between cells. Clustering multiple data streams concurrently has been done using a ClusTree algorithm [16,17]. Synopses of different concurrent streams are kept up in an index structure depend on R-tree. Keeping up summary statistics of each data object leads to a larger workload. When new data comes, the algorithm looks for the closest micro-cluster by navigating the tree of which leaf nodes have all the micro-clusters. The greatest hindrances of this algorithm are squandering more memory [17]. The addition of the conventional clustering algorithms Neural Gas (NG) and the Self-Organizing Map (SOM) for clustering data streams are proposed [19]. The extension is reliant on a simple patch decomposition of the data set and only requires a fixed memory space.
For most of the existing clustering parallel implementations, the clustering quality is commonly less accurate than the sequential implementation [1]. This is because the clustering quality is affected by the methods utilized in the data partitioning and consolidating phase. A common problem as well is that algorithms can’t cope efficiently with fast-evolving data streams and consume large memory for tracking the clusters. For most of the presented related works, the processing time has been regarded as the major performance measure to be tracked, with no additional measures, such as the clustering quality and the scheduling delay.
3. Spark Streaming architecture
The principle concept in spark streaming is a discretized stream (DStream) [22, 23] which is a consecutive sequence of distributed collections of elements describing a continuous stream of data, and which is called RDDs (Resilient Distributed Datasets). DStreams can be created in two ways; either from a source (e.g. data from a socket, Kafka, file stream, etc.) or by transforming current DStreams utilizing parallel operators (e.g. Map, Reduce, and Window). RDDs are usually, partitioned across multi-cores by Spark. In spark, all the data is represented as RDDs and all DStream operations as RDD operations [24,25, 26].
The number of RDDs partitions created can be specified. The stream processing model in spark is a micro-batch processing. Data received by Input DStreams are processed using DStream operations.
The Spark cluster is responsible for scheduling and dividing the resource in the machine. The main target of the cluster manager is to divide the applications across resources to run the application in parallel mode. Apache Spark has three kinds of cluster managers, standalone, Hadoop YARN, and Apache Mesos. In this work, a standalone cluster manager is utilized.
In the view of master-slave architecture, Apache Spark has one master process and enormous worker processes. These are the following: – the master process consists of a job tracker and a name node. The job tracker is responsible for scheduling jobs and assigning the jobs to the task tracker on the worker process, which is responsible for executing the map and reduce function. The name node is responsible for the storage and management of file metadata and file distribution across several data nodes on worker nodes, which contain the data contents of these files.
Spark gives a graphical UI (user interface) to following the performance of applications. The two important metrics in web UI are 1) Scheduling Delay – the time a batch remains in a queue for the processing of prior batches to end. 2) Processing Time – the utilized time to process every batch of data. The stream processing model in spark is micro-batch processing, processing one batch at a time, so batches wait in the queue until the prior batches finish. The mechanism for minimizing the processing time of each batch and scheduling delay is to increase the parallelism [24, 25].
4. Parallel clustering approach for data streams
Parallel processing provides an optimized solution for clustering huge data streams given that the accuracy of clustering is preserved. In this section, the proposed parallel design of the SCluStream algorithm is described.
4.1. SCluStream
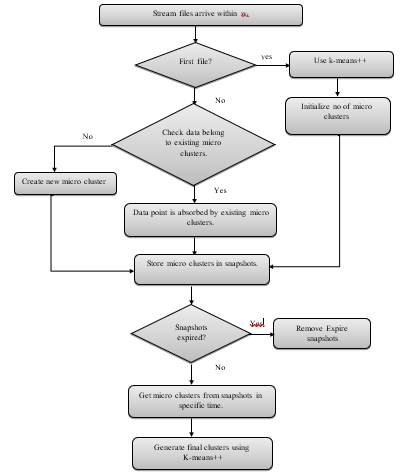
SCluStream saves more time and memory by processing the most recent transactions that fall within window size and the old data are eliminated, so SCluStream overcomes the main obstacles in data streams: – time and memory resources. In the proposed approach, SClustream is implemented on parallel processing utilizing a multicore platform to increase the performance of handling the large volumes of data streams. Figure 1 shows the primary steps of SCluStream and the connections between these steps. The representation of sequence steps and processes is described by the flowchart shown in figure 2. SCluStream consists of three phases.
1) Online phase
- Toward the start of the algorithm implementation, the q initial micro-clusters are generated from the incoming real-time data by applying k-means++ within the time window .
- The window time is kept up for following the latest instances falling during the window size of the data stream and dispensing the outdated data.
- Keeping the statistical data instead of keeping all incoming data in micro-clusters.
- A micro-cluster comprises of the following components {N, LS, SS, LST, SST} where, N the data points number, LS the linear sum of the N, and SS the squared sum of the N. The two last LST and SST are the sum and the sum of the squares of the timestamps of the N.
- When data point arrival, a single of the subsequent two possibilities take place: –
- i) The data point is consumed by one of the current micro-clusters. This assimilation is based on the nearness of the cluster to the data point; this is determined by applying a distance metric among the centroid of the micro-cluster and the data point. So, the data point is consumed to the closest micro cluster.
- ii) The data point is put in its specific new micro-cluster, however with the restriction that the micro-clusters number stays fixed. Thus, the current clusters number should be decreased by one, which can be accomplished by merging the two nearest micro clusters jointly.
2) Expiring phase
The snapshots are used for storing micro-clusters at each time slot t within a time window . The establishment time for those expiring snapshots doesn’t lie among the current time and the window time subtracted from current time. Defining the expiring snapshots from < – where is the establishment time of snapshot, is the current clusters number time, is the window time.
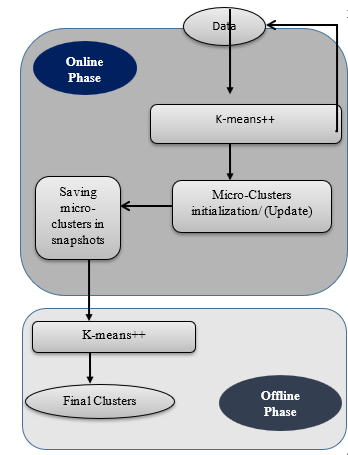 Figure 1: The primary steps of SCluStream [8]
Figure 1: The primary steps of SCluStream [8]
3) Offline phase
The offline phase is consolidating the micro-clusters to deduce the final clusters. The offline phase obtains the micro clusters from kept snapshots during the time window wt. The final macro clusters are defined by utilizing k-means++ rather than conventional k-means to get more accurate results.
4.2. Parallel SCluStream processing
Partitioning the input data streams into a number of partitions. Perform Parallel processing of k-means++ algorithm in online phase on each partition to create micro clusters. Expanding the number of parallel tasks that one executor can execute by expanding the number of available cores to execute per executor. The number of partitions input data streams and operations are reliant on the existing number of cores. Each core is responsible for processing one partition. The steps in figure 3 show the primary steps of parallel-SCluStream and the connections among these steps.
The Steps of SCluStream algorithm involved in the parallel processing can be listed as follow:
- In the initialization phase, the data is split into p
- The online phase parallel processing can be described in the following steps.
- The distance calculation from every data point x to every centroid by utilizing Euclidean distance, the distance between every data point x and the c
D (?, ć) = (1)
- This step can be parallelized due to the volume of time used for parallelizing this step brings, an advantage by reducing the total processing time required for finishing this step as multiple cores will process some chunks of the complete dataset simultaneously.
- Assign data point x to the nearest centroid reliant on distance calculation in the previous step, this process requires a number of comparisons (reliant on the number of the clusters required to be obtained) for choosing the cluster where the data point x should be included. The comparison process will execute in multiple threads, every thread will assign only part of the records of the entire dataset to a single cluster.
- Compute a set of new partial centroids for each of the processed partitions. The distance between the new centroids is calculated.
- Compute new centroids for the whole data set and match the values of the new centroids, with the values of the centroids at the aforementioned iteration. If the new centroids are different from the previous iteration go to setp2; else continue to generate final clusters.
If the number of micro clusters is obtained then the final micro clusters is generated; else merging two nearest micro-clusters by using additive property of micro-cluster [7] data structure to generate the determined number of final clusters reliant on the distance calculated in step 3. The additive property means, the
nearest micro-clusters can be consolidated into a new micro-cluster by merging their components [7, 8]. The additive property for merging two nearest micro-clusters is declared in the following equations:
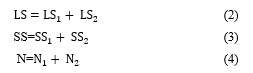
 5. Experimental study
5. Experimental study
5.1. Experimental setup
All experiments have been implemented on a standalone implementation, where master process and worker processes all reside on a single machine. The machine used is a laptop with a processor core I5, 8G memory, and an Ubuntu 64-bit operating system. The cluster processing framework used is an apache spark using scale 2.10. The proposed approach has been tested on two datasets: one small and another large one. The results on both datasets show significant improvements in the efficiency of clustering in terms of processing time and scheduling delay without diminishing the clustering quality. The small and large datasets used for testing are respectively: 1) KDD-CUP’99, consisting of 494,021 rows and 43 attributes, and 2) KDD-CUP’98, consisting of 95,412 rows and 56 attributes. For the execution of the framework, configurations are set at first. The parameter settings identified in table 1 have been identified for experimentation.
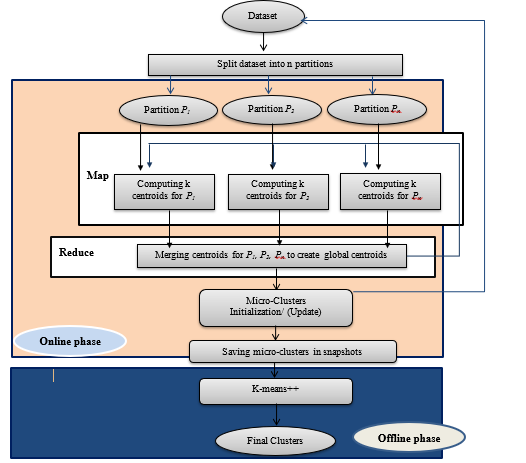 Figure 3: The primary steps of Parallel-SCluStream
Figure 3: The primary steps of Parallel-SCluStream
Table 1: Parameter Settings Utilized in the Experimentation
| Parameters | Value |
| The date points number n | 2000 |
| The micro clusters q number | 50 |
| The dimensions number in KDD-CUP’99 data set d | 31 |
| The dimensions number in KDD-CUP’98 data set d | 56 |
| Window size wt | 70s |
| The cores’ number | 3 |
In the settings, the cores’ number is set to 3 cores because the laptop utilized in implementation consists of four cores, one core for the operating system and 3 cores for running the application. This setting is done in spark configuration to determine the maximum available cores for running applications on parallel processing. In the experimental study of the non-parallel SCluStream implementation, the clustering quality was tested with the different number of micro clusters and it was better when the number of the cluster was 50. Also, the clustering quality was tested at different window sizes of and it was better when the size of the window was equal to 70 [8]. Therefore, we used this number of micro clusters and the size of the window in the parameters setting for the parallel SCluStream implementation. The number of initial points can change (up or down), provided that it does not exceed the size of the streamed files.
For assessing the performance of the parallel architecture shown in figure 3, the following measures are recorded:
1) Scheduling Delay TSD – the time a batch remains in a queue for the processing of prior batches to end.
2) Processing Time TP – the utilized time to process no of the batches of data.
3) Clustering Quality (SSQ) – SSQ is measured for sequential processing and parallel processing. The distance D among data point and the closest centroid is calculated . The SSQ is calculated as the sum of for the whole points in current window. The smaller the value of SSQ, the superior the clustering quality.
![]() The dataset is divided into p partitions, where each partition has been processed in a separate core. The partitioning process is done in the initialization phase. In the map phase, every partition is processed in an isolated core to generate partial clusters. In the reduce phase merge partial clusters in one core to create final clusters.
The dataset is divided into p partitions, where each partition has been processed in a separate core. The partitioning process is done in the initialization phase. In the map phase, every partition is processed in an isolated core to generate partial clusters. In the reduce phase merge partial clusters in one core to create final clusters.
5.2. The experimental results
1) Scheduling Delay
Figure 4 shows the scheduling delay of SCluStream for sequential and parallel processing for the different file sizes in the two real datasets. The average scheduling delay decrease with increment the level of parallelism by increment the number of partitions for input data and operations. But when the number of the partitions is bigger than the available cores number, the scheduling delay increment. When the number of partitions is five so 3 partitions are processing in parallel and the 4th, 5th partition waits until one of three cores is workless. The results proved that the average scheduling delay decreases by increase the parallelism but with restriction to an available number of cores in the machine.
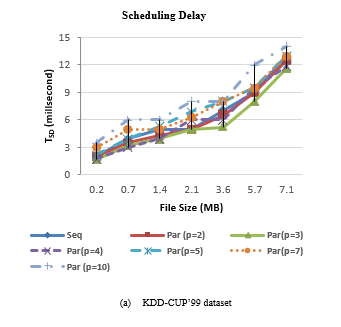
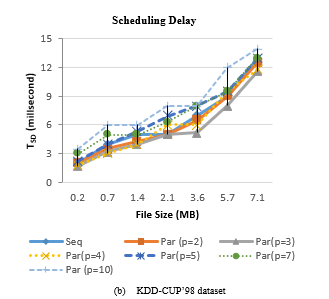 Figure 4: Scheduling delay of SCluStream for sequential and parallel processing for different file size
Figure 4: Scheduling delay of SCluStream for sequential and parallel processing for different file size
2) Processing time
Figure 5 illustrates the processing time for sequential and parallel processing of SCluStream versus different files size in the two real datasets. In figure 5-(a) sequential processing of 7.1MB file size which consists of 100000 records by SCluStream takes 13.2 seconds for KDD-CUP’99 data set. Processing 100000 points by parallel processing of SCluStream takes around 11 second when repartition the input data streams to 2 partitions, every partition being processed in a separate core simultaneously. Processing 100000 points takes around 9.6 second when repartition the input data streams to 3 partitions. The processing time decrease when the number of partitions is less than or equal to the total number of cores as illustrated in figure 5 when repartition the input data streams to 2 or 3 partitions.
The 3 partition processes will execute in parallel reliant on available cores and the partition process will process when one of the 3 cores, is workless. The average processing time for processing different bathes of data when repartition the input data to 5 partitions is nearly equal to the average processing time for sequence processing.
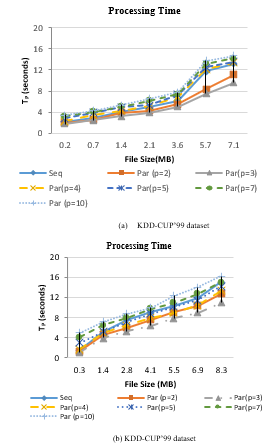 Figure 5: Processing time of SCluStream for sequential and parallel processing for different file size.
Figure 5: Processing time of SCluStream for sequential and parallel processing for different file size.
In figure 5-(b) sequential processing 8.3MB file size which consists of 90000 records by SCluStream requires 15 seconds for KDD-CUP’98 data set. Processing 90000 points by parallel processing of SCluStream requires around 12.8 second when repartition the input data streams and to 2 partitions and takes around 11 second when repartition the input data streams and operations to 3 partitions. The processing time increase about 9.4 seconds from 4 partitions to 3 partitions and 3.4 seconds from the 4 partitions to 2 partitions because the 3 partition processes will run in parallel as there are three cores and the 4th partition process will process when one of the 3 cores, is workless. When the number of partitions is set to 5 partitions, the average processing time is nearly less than the average processing time for Sequential processing for different bathes of data in figure 5-(b) by 0.4 second. The average processing time for processing different bathes of data when repartition the input data to 7 and 10 is bigger than the average processing time for sequence processing. Assuming the dataset is part in numerous parts, not all parts will be handled in a similar time since the number of cores that will execute on a machine is bigger than the maximum number of cores that can be processed by that machine, implying that some of the cores will stand by till they have access to the CPU.
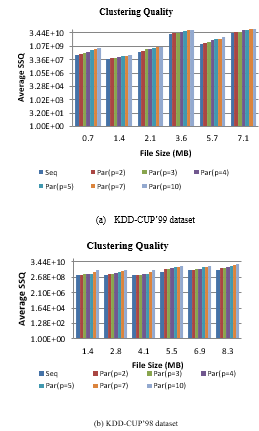 Figure 6: Clustering Quality (SSQ) of sequential and parallel SCluStream for different file size
Figure 6: Clustering Quality (SSQ) of sequential and parallel SCluStream for different file size
3) Clustering quality
Figure 6 illustrates the comparison among the clustering quality (SSQ) for sequence and parallel processing of SCluStream versus different files size at the same parameters in experimental configuration (points number 2000, micro clusters number (q=50), window time (70 s)) in the two real datasets. Figure 6-(a) obviously shows that the average SSQ of sequential-SCluStream is approximately close or equal to the average SSQ of parallel SCluStream at a different level of parallelism, especially from partition 2 to partition 3 but from partition 4 to partition 10 the average SSQ is slightly affected from the average SSQ of sequential-SCluStream.
The average SSQ of parallel-SCluStream, especially from partition 2 to partition 4 is nearly close to the average SSQ of sequential-SCluStream in figure 6-(b) and the average SSQ of parallel-SCluStream from partition 5 to partition 10 is slightly affected from the average SSQ of sequential-SCluStream.
5.3. Results Discussion
The experimental results have presented that the parallel implementation for the SClustream managed to superiority the sequential SCluStream for the different parameter studied settings when testing on the two real datasets. Experiments for the processing time have shown that the parallel implementation for the SClustream is nearly 1.5 times quicker than the sequential SCluStream for both datasets. Experiments for the scheduling delay time have shown that the SClustream parallel implementation is approximately between 1.3 to 1.7 times less than the sequential SCluStream for both datasets. Scalability levels of the proposed approach were evaluated by varying the file sizes for the two datasets at the same number of cores, and results have shown the proposed approach succeeds to optimize the processing time and scheduling delay while remaining the clustering quality near or identical to the sequential SCluStream.
In the experimental setting, the number of cores is set to 3 cores according to the capability of the available computer. The data set has been divided the data set to 3 partitions suitable to the number of cores, making every core is responsible for processing one data partition. In this case, the proposed approach succeeded in decreasing the processing time of the dataset and decreasing the scheduling delay for every batch waiting for the previous batch. In case the dataset is divided into bigger than 3 partitions for the current implementation setup, then not all partitions would have been processed simultaneously, because the number of partitions running on a machine is bigger than the maximum number of cores for our used implementation. Hence, some of the partitions will need to wait until one of the cores become workless. In such case, the processing time and scheduling delay will probably bigger than the processing time and scheduling delay of the sequential execution. Running a cluster implementation, instead of the standalone implementation, is expected to provide a promising solution in regard to maintaining the clustering quality obtained, and with much more speed factor than the obtained in the standalone implementation. The conclusive setting for the standalone implementation is that for ensuring the best running parallel mechanism, it is recommended to set the number of partitions equal to the cores’s number in the node so that all the partitions will process in parallel and the available resources will be utilized optimally.
6. Conclusion and future work
Recently, data stream clustering is becoming vital research. This problem needs a process capable of clustering continuous data while considering the constraints of memory and time and generating clusters with high quality. In this paper, parallel clustering implementation on MapReduce and apache spark framework is for the clustering algorithm (SCluStream), which is an efficient algorithm for tracking clusters over sliding window mechanism, focusing on the latest transactions to speed up processing and execution. The implementation has been presented on a standalone cluster manager. The experimental study proved that the parallel standalone implementation with the multi-core processing is successful to take less processing time by approximately 1.5 times and between 1.3 to 1.7 times less scheduling delays than the non-parallel SCluStream implementation. Regarding the clustering quality, it is approximately equal to that of the non-parallel implementation. For obtaining much more clustering algorithm acceleration future work will consider the implementation in connected nodes by using a spark cluster, master nodes, and many worker nodes while making the best configuration and utilization of available executors and cores. Also, apply the best mechanisms for data partitioning and distribution. The automatic determination for all parameter settings will be applied in future work. In addition to comparing the parallel implementation of SCluStream with other data streaming parallel clustering algorithms such as pGrid with various real data sets to confirm the quality and performance for the parallel implementation of SCluStream compare to other algorithms.
- C.Liu, R.Ranjan, X.Zhang, C.Yang, D.Georgakopoulos, and J.Chen. “Public auditing for big data storage in cloud computing–a survey”. Proceedings – 16th IEEE International Conference on Computational Science and Engineering, CSE 2013, 1128–1135. doi: 10.1109/CSE.2013.164.
- B.Val, Pablo, N.F.Garcia, L.S.Fernández, and J.A.Fisteus. “Patterns for distributed real-time stream processing”. IEEE Transactions on Parallel and Distributed Systems, 28(11), 3243–3257, 2017, doi.org/10.1109/TPDS.2017.2716929.
- N.Kaur, and S.K.Sood. “Efficient resource management system based on 4vs of big data streams”. Big data research, 9, 98-106, 2017, doi.org/10.1016/j.bdr.2017.02.002.
- T.S.Sliwinski, and S.L.Kang. “Applying parallel computing techniques to analyze terabyte atmospheric boundary layer model outputs”. Big Data Research, 7, 31–41, 2017, doi: 10.1016/j.bdr.2017.01.001.
- I.I.Yusuf, I.E. Thomas, M.Spichkova, and H.W. Schmidt. “Chiminey: Connecting scientists to hpc, cloud and big data”. Big Data Research, 8, 39–49., 2017, doi: 10.1016/j.bdr.2017.01.001.
- Z.Lv, H.Song, P.B.Val, A.Steed, and M.Jo. “Next-generation big data analytics: State of the art, challenges, and future research topics”. IEEE Transactions on Industrial Informatics, 13(4), 1891–1899, 2017, doi: 10.1109/TII.2017.2650204.
- J.A.Silva, E.R.Faria, R.C. Barros, E.R. Hruschka, A.C.d.Carvalho, and J.Gama. “Data stream clustering: A survey”. ACM Computing Surveys (CSUR), 46(1), 1–31, 2013, doi: 10.1145/2522968.2522981.
- D.Sayed, S.Rady, and M. Aref. “SCluStream: an efficient algorithm for tracking clusters over sliding window in big data streaming”. International Journal of Intelligent Computing and Information Sciences, 19(2), 1–19., 2019, doi:10.21608/IJICIS.2019.62592.
- C.C.Aggarwal. “Data streams: models and algorithms”. Springer Science & Business Media, vol. 31, 2007, doi: 10.1007/978-0-387-47534-9.
- A. Bousbaci, and N.Kamel. “Efficient data distribution and results merging for parallel data clustering in map reduce environment”. Applied Intelligence, 48(8), 2408–2428.
- M.M.Patwary,A.D.Palsetia, A.Agrawal, W.k.Liao, F.Manne, and A.Choudhary. “A new scalable parallel DBSCAN algorithm using the disjoint-set data structure”. SC’12: Proceedings of the International Conference on High Performance Computing, Networking, Storage and Analysis, 1–11, 2012, doi: 10.1109/SC.2012.9.
- T. Sakai, K.Tamura, K.Misaki, and H.Kitakami. “Parallel processing for density-based spatial clustering algorithm using complex grid partitioning and its performance evaluation”. Proceedings of the International Conference on Parallel and Distributed Processing Techniques and Applications (PDPTA), 337, 2016, doi:10.1109/TPDS.2019.2896143.
- Y. He, H.Tan, W.Luo, S.Feng, and J.Fan. “MR-DBSCAN: a scalable MapReduce-based DBSCAN algorithm for heavily skewed data”. Frontiers of Computer Science, 8(1), 83–99., 2014, doi: 10.1007/s11704-013-3158-3.
- X.Sun, and Y.C.Jiao. “pGrid: Parallel grid-based data stream clustering with mapreduce”. Report. Oak Ridge National Laboratory. 2009.
- Y. Gong, R.O.Sinnott, and P.Rimba. “Rt-dbscan: Real-time parallel clustering of spatio-temporal data using spark-streaming”. International Conference on Computational Science, 524–539, 2018, doi: 10.1007/978-3-319-93698-7.
- P.Kranen, I.Assent, C.Baldauf, and T.Seidl. “The ClusTree: indexing micro-clusters for anytime stream mining”. Knowledge and Information Systems, 29(2), 249–272, 2011, doi: 10.1007/s10115-010-0342-8.
- Z.R.Hesabi, T.Sellis, and X.Zhang. “Anytime concurrent clustering of multiple streams with an indexing tree”. Workshop on Big Data, Streams and Heterogeneous Source Mining: Algorithms, Systems, Programming Models and Applications, 19–32, 2015, doi: 10.1.1.1080.3236.
- R.Tashvighi and A.Bagheri. “PPreDeConStream: A Parallel Version of PreDeConStream Algorithm”. International Journal of Computer Applications, 975, 8887, 2016, doi: 10.5120/ijca2016912235.
- N. Alex, A.Hasenfuss, and B.Hammer. “Patch clustering for massive data sets”. Neurocomputing, 72(7–9), 1455–1469, 2009, doi: 10.1016/j.neucom.2008.12.026.
- G.Mencagli, D.B.Heras, V.Cardellini, E.Casalicchio, E.Jeannot, F.Wolf, A.Salis. “Euro-Par 2018: Parallel Processing Workshops: Euro-Par 2018 International Workshops”. In 24th International Conference on Parallel and Distributed Computing, Euro-Par 2018 Turin, Italy. Vol. 11339. Springer, 2018.
- I.D.Borlea, R.E.Precup, F.Dragan, A.B.Borlea. “Parallel Implementation of K-Means Algorithm Using MapReduce Approach”. In IEEE 12th International Symposium on Applied Computational Intelligence and Informatics (SACI), 2018, doi: 10.1109/SACI.2018.8441018.
- M. Zaharia, M. Chowdhury, M. J. Franklin, S. Shenker, and I. Stoica. “Spark: Cluster computing with working sets”. HotCloud, 10(10–10), 95, 2010.
- Z.Yunquan, T.Cao, Shigang Li, Xinhui Tian, L.Yuan, H.Jia, and A.V. Vasilakos. “Parallel processing systems for big data: a survey”. Proceedings of the IEEE, 104(11), 2114–2136, 2016, doi:10.1109/JPROC.2016.2591592.
- K.Holden, A.Konwinski, P.Wendell, and M.Zaharia. “Learning spark: lightning-fast big data analysis”. O’Reilly Media, Inc., 2015.
- M. Zaharia, M. Chowdhury, M. J. Franklin, S. Shenker, and I. Stoica. “Spark: Cluster computing with working sets”. HotCloud, 10(10–10), 95, 2010.
- V.Sanz. Marco, B.Taylor, B.Porter, and Z.Wang. “Improving spark application throughput via memory aware task co-location: A mixture of experts approach”. Proceedings of the 18th ACM/IFIP/USENIX Middleware Conference, 95–108. 2017, doi: 10.1145/3135974.3135984.